L
Lord Cola
Guest
Abstract
Background
Coronavirus Disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has now been confirmed worldwide. Yet, COVID-19 is strangely and tragically selective. Morbidity and mortality due to COVID19 rise dramatically with age and co-existing health conditions, including cancer and cardiovascular diseases. Human genetic factors may contribute to the extremely high transmissibility of SARS-CoV-2 and to the relentlessly progressive disease observed in a small but significant proportion of infected individuals, but these factors are largely unknown.Main body
In this study, we investigated genetic susceptibility to COVID-19 by examining DNA polymorphisms in ACE2 and TMPRSS2 (two key host factors of SARS-CoV-2) from ~ 81,000 human genomes. We found unique genetic susceptibility across different populations in ACE2 and TMPRSS2. Specifically, ACE2 polymorphisms were found to be associated with cardiovascular and pulmonary conditions by altering the angiotensinogen-ACE2 interactions, such as p.Arg514Gly in the African/African-American population. Unique but prevalent polymorphisms (including p.Val160Met (rs12329760), an expression quantitative trait locus (eQTL)) in TMPRSS2, offer potential explanations for differential genetic susceptibility to COVID-19 as well as for risk factors, including those with cancer and the high-risk group of male patients. We further discussed that polymorphisms in ACE2 or TMPRSS2 could guide effective treatments (i.e., hydroxychloroquine and camostat) for COVID-19.Conclusion
This study suggested that ACE2 or TMPRSS2 DNA polymorphisms were likely associated with genetic susceptibility of COVID-19, which calls for a human genetics initiative for fighting the COVID-19 pandemic.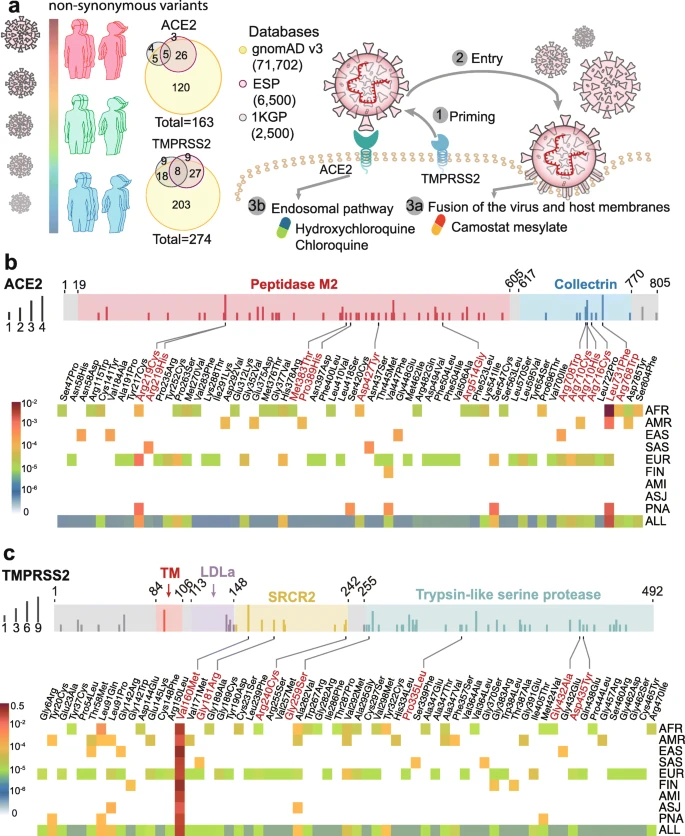
The coding-region variants in ACE2 and TMPRSS2 from ~ 81,000 human genomes across 8 populations. a Coding-region variants in the genes encoding angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) across three human genome databases: (i) Genome Aggregation Database (gnomAD v3), (ii) Exome Sequencing Project (ESP), and (iii) 1000 Genomes Project (1KGP). SARS-CoV-2 utilizes the host cell factors angiotensin-converting enzyme 2 (ACE2) for entry into cells and the host transmembrane serine protease TMPRSS2 for SARS-CoV-2 spike (S) protein priming, offering potential pathway for therapeutic development in treatment of COVID-19. b Distribution of 61 deleterious variants in the ACE2 coding region identified in gnomAD (v3). Polyphen2 > 0.96 and CADD scores > 20 as cutoff identify putative deleterious variants. The upper panel using 3 colors shows the functional domains of ACE2, and the height of the vertical line represents the number of populations that carry this variant. The lower heatmap shows the allele frequencies (color key) of a variant across different populations. c Distributions of 63 putative deleterious variants in the TMPRSS2 coding region using the same approach of b. AFR, African/African-American; AMI, Amish; AMR, Latino/Admixed American; ASJ, Ashkenazi Jewish; EAS, East Asian; FIN, Finnish; EUR, Non-Finnish European; SAS, South Asian; PNA, population not assigned
Full article: New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis - BMC Medicine