Travis
Member
- Joined
- Jul 14, 2016
- Messages
- 3,189
DNA Methylation: The Confusion over 5-Methylcytosine
The methylation of DNA is an undeniable phenomenon, and found in most living organisms. The enzymes responsible for this have been extracted, purified, assayed—with their kinetic rates determined, giving indisputable proof that DNA methylation can be an enzymatic event. The DNA encoding for such enzymes have been genetically sequenced, synthesized, and recombined to form fully-functional constructs . . . and the X-structures have been elucidated.
But there exists three forms of methylated DNA: oxygen-linked, as in the case of 6-O-methylguanidine; nitrogen-linked as in the case of N-6-methyladenosine, N-3-methylthymine, and N-4-methylcytosine; and surprisingly, carbon-linked as in the single case of C-5-methylcytosine. All of these modified nucleotides exist and can be found in DNA, and one of them has dubious origins—a biolochemical unicorn.

While this may seem trivial, it is not; as you will see. Misunderstandings surrounding the origin of 5-methylcytosine obfuscates epidemiological correlations, hides important cellular control mechanisms, and can perhaps can even lead to further misunderstandings if compounded—snowballed around the original misconception.
I. The Modern Paradigm
But a clear trend has emerged: Methylated DNA strands appear to be more stable; they are slighly-downregulated genes. The methylation of DNA is the protoypical epigenetic mechanism, and the only one acknowledged by many biologists.
These methylations are also thought to protect cellular DNA from its own enzymes—enzymes which could exist to destroy foreign DNA, or as simply a recycling measure. There are many other speculations on the function and purpose of DNA methylation, but a few things should perhaps be considered before entertaining some of the more abstract and tenable hypotheses out there.
Jeltsch, Albert. "Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases." Chembiochem (2002)
II. Why Carbon:Carbon Bonds are Different
The formation of the carbon:carbon bond is a relatively rare event in biochemistry—almost unheard-of without a carbonyl or phosphate involved. The electronegative oxygen pulls electrons from the carbonyl carbon, allowing backside attack from electron-rich nucleophiles such as a thiols and amides. The condensation reactions in steroid synthesis make use of phosphate groups on the terminal carbons, 'activating' them with electronegativity.
The DNA methyltransferase reaction—which catalyzes the carbon:carbon bond formation of 5-methylcytosine—involves adding a methyl group to a resonant ring. You'd be hard-pressed to find another instance of such, or even another biochemical carbon:carbon bond formation besides maybe the rare Diels–Alder.
The #5 bond of methylcytosine is the only variable carbon:carbon bond in DNA. All other methylations are to either to nitrogen or oxygen, additions which are in no way unusual: Such examples can be found in familiar molecules such as melatonin, methoxytyramine, and epinephrine. So when talking about 'DNA methylation' it becomes necessary to make the distinction between what atoms have been methylated, or assumed to have been.. .
"So far, in metazoa only cytosine-C5 methylation has been found in DNA; this methylation mainly occurs at CG sequences, about 60 ± 90% of which are modified in mammals (corresponding to 3 ± 8% of all cytosine residues)." —Albert Jeltsch
The methylated DNA base called 5-methylcytosine gets the most attention, as it's known to be the most effective at silencing genes—affecting both double-helix stability and replication more than the N-linked and O-linked varieties.
It was also the first one discovered.
Jeltsch, Albert. "Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases." Chembiochem (2002).
III. Help from DNA Methyltransferase
"Therefore, the catalytic mechanism of C-MTases can explain why DNA MTases developed a base-flipping mechanism." ―Jeltsch
In step four, an unidentified base materializes out of nowhere to abstract a proton from carbon five.
This is a base later found to not exist at 2.8 Å resolution.(Reinisch, 1995).

Of course all of this is too small to see, so they can just sprinkle fairy dust to obscure any irregularities. Step four is technically the molecular equivalent of Bruce Lee in "The Way of the Dragon."
"Although MET supplementation significantly decreased the [SAM]:[SAH] ratio in liver and brain, no significant dietary effects on genome-wide DNA methylation were found." —Robert A. Waterland
All so-called DNA methyltransferase enzymes use S-adenosylmethionine as cofactor. One would then expect this cofactor to increase total DNA methylation by ingestion or by injection. It does, after all, the source of the methyl groups in question.
The failure of studies to demonstrate changes in 'DNA methylation'—as defined as 5-methylcytosine—by methionine supplementation shouldn't go unnoticed. It's unfortunate that many researchers fail to differentiate between the possible and the impossible methylation events in titles and abstracts, as this only adds confusion to an already confused paradigm.
Waterland, Robert A. "Assessing the effects of high methionine intake on DNA methylation." The Journal of nutrition (2006)
Do Amaral, Cátia Lira. "The effects of dietary supplementation of methionine on genomic stability and p53 gene promoter methylation in rats." Mutation Research/Genetic Toxicology and Environmental Mutagenesis (2011)
V. Failure of Folate to do Likewise
Although not considered a cofactor for the DNA methyltransferase reactions, folate is dragged into this imply because it regenerates S-adenosylmethionine. But if S-adenosylmethionine itself cannot even influence DNA methylation, how can you folate capable of this?
When measuring the effects of folate on hundreds of people, the results were negligible—basically null.
"Women with higher (vs. lower) RBC folate had higher mean DNA methylation (5.12 vs. 4.99%) in the pre-fortification period, but lower (4.95 vs. 5.16%) DNA methylation in the post-fortification period." —Sajin Bae
This was defined with respect to 5-methylcytosine, or course. Increases in nitrogen-methylated DNA is certainly possible from supplemental methionine, but N-methylation is rarely studied.
Should anyone think that a .03% change is something to consider, take a look at what vitamin B₁₂ can do. It can cause great changes in spite of the fact that it's not a cofactor for DNA methyltransferse.
Crider, Krista S. "Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role." Advances in Nutrition: An International Review Journal (2012)
Bae, Sajin, et al. "Impact of folic acid fortification on global DNA methylation and one-carbon biomarkers in the Women's Health Initiative Observational Study cohort." Epigenetics (2014)
VI. Vitamin B₁₂ Increases Methylation—and Nothing Else
By unambiguous methods: Vitamin B₁₂ had been shown to increase cytosine methylation; something which cannot be achieved reliably by either folic acid or S-adenosylmethionine—the definitive methyl donor, making this especially enigmatic since S-adenosylmethionine is the very cofactor for the enzyme thought responsible.
Choi, Sang-Woon, et al. "Vitamin B-12 deficiency induces anomalies of base substitution and methylation in the DNA of rat colonic epithelium." The Journal of nutrition (2004)
VII. How 5-Methylcytosine is Actually Formed
Regular cytosine is formed from the amino acid aspartate. This is a straghtforward process, and had been known since the '50s.
A small metabolite called carbamoyl phosphate is attacked by the amide (shown below as an amine but the hydrogen is removed by the enzyme) of aspartate, held in position and acted-upon by the enzyme carbamoyl transferase. This is not a carbon:carbon addition, and neither is the final condensation stage. Such interactions are known to occur all the time.

But when β-methylaspartate is acted upon by these enzymes, you would expect the 5-methylcytosine precursor to be formed: 5-methyldihydroororate, or 5-methyldihydroororic acid (depending on pH.)

Which would then go on to become 5-methylcytidine, and then incorporated into DNA. The substrate β-methylaspartate is no stranger to biology: This molecule is formed through the vitamin B₁₂-catalyzed enzyme glutamate mutase—named so because it actually isomerizes glutamate into β-methylaspartate, similar to B₁₂'s ability to isomerize methylmalonic acid.
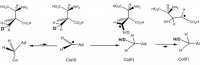 Vitamin B₁₂ is only thing capable of isomerizing glutamate into β-methylaspartate.
Vitamin B₁₂ is only thing capable of isomerizing glutamate into β-methylaspartate.
Now this is the formation of a carbon:carbon bond, to be sure, and could appear no less difficult than the one in question: the methlyation of cytosine on single-stranded DNA. But keep-in-mind that vitamin B₁₂, or cobalamin, is a giant heme-like ring structure with a cobalt atom in the centre. This gives it high electric potential, something that the DNA methyltransferase or S-adenosylmethionine simply doesn't have.
When the glutamate/aspartate ratio is high, you might expect more glutamate to be be converted to β-methylaspartate through the action of glutamate mutase. This is the precursor the 5-methylcytosine, and has nothing do with methionine.
Yates, Richard A. "Pyrimidine biosynthesis in Escherichia coli." Journal of Biological Chemistry (1956)
VIII. Confirmation from Neurotransmitter Studies
You might also expect this to happen anywhere . . . even in the brain, the place most suitable for such an investigation. The brain has a highly-variable glutamate/aspartate ratio, and thus is a prime candidate for such analysis. These two amino acids are considered to be neurotransmitters involved in learning, motivation, and memory.
Following in the footsteps of Francis Crick, Jarrod Meadows had decided to investigate the effect of glutamate on DNA methylation in the brain. He used tetrodotoxin, as this is the standard molecule used to inhibit glutamate release in the brain. The low brain glutamate was confirmed, and he had found that this had resulted in DNA methylation changes greater than those resulting from inhibiting DNA methyltransferase. Tetrodotoxin, ostensibly through glutamate, did this in spite of contributing no methyl groups whatsoever.
Decreasing glutamate would inhibit the formation of β-methylaspartate, the precursor for 5-methylcytosine, through glutamate mutase. This would lead to lower DNA methylation, exactly as was found.
Other studies have confirmed such a relationship, between asparate levels and DNA methylation (Punzo, 2016).
In the Meadow's study: The genes hypomethylated from low glutamate were, and perhaps not surprisingly, involved in aspartate signalling:
This strengthens the idea that the glutamate/aspartate ratio is controlling DNA 'methylation' on the very genes responsible for its regulation—having nothing to do with methionine. This might seem paradoxical, if not for the facts outlined above. With these in mind, this all makes perfect sense.
Meadows, Jarrod P., et al. "DNA methylation regulates neuronal glutamatergic synaptic scaling." Science signaling (2015)
Punzo, Daniela, et al. "Age-related changes in D-aspartate oxidase promoter methylation control..." Journal of Neuroscience (2016)
IX. Hints from Genetics
The methylation of DNA—as defined by 5-methylcytosine—is routinely measured on select genes. This is because: Some regions are particularly-rich in cytosine, leading to more accurate measurements in these places. One of the genes commonly tested for methylation happens to be the one encoding for D-aspartate oxidase:
You would then think that highly-'methylated' areas would be found in the genes responsible for controlling glutamate:aspartate metabolism, including—but not limited to—aspartate transaminase, γ-glutamyl transferase, and glutamyl mutase.
Florio, Ermanno, et al. "Tracking the evolution of epialleles during neural differentiation and brain development: D-Aspartate oxidase as a model gene." Epigenetics (2017)
X. Five-Methycytosine
Was the sixth nucleotide discovered, and the first assumed to be a post-translational modification. Originally discovererd in 1925, it's existence hadn't been confirmed until 1948. Two years later its existence was headlined in the high-visibility journal Nature.
"...nevertheless, the fraction is distinct from cytosine and is clearly not uracil." ―Rollin D Hotchkiss
Five-methylcytosine was on the minds of researchers then, and the nitrogen-methylated nucleotides weren't known to exist until later.
Johnson & Coghill. "Researches on pyrimidines. The discovery of 5-methyl-cytosine in tuberculinic acid..." Journal of the American Chemical Society (1925)
Hotchkiss, Rollin D. "The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography." Journal of Biological Chemistry (1948)
Wyatt, G. R. "Occurrence of 5-methyl-cytosine in nucleic acids." Nature (1950)
XI. DNA Methyltransferase
With 5-methylcytosine the only known 'DNA methylation' product, any transfer of methyl groups measured from S-adenosylmethionine to DNA would simply been assumed to be those forming 5-methylcytosine. The variable nature of 5-methylcytosine content found in DNA further would further confound such measurements, turning the negligible enzymatic rates into something even less.
Moreover, Sneider was likely measuring the sum of all of the DNA methytransferase rates combined, even the real ones. This is a possibility that he'd even mentioned himself, a likelihood which would further marginalize the already snail-like catalytic rates. He also didn't prove that the methyl groups were actually transferred to cytosine at carbon five; Sneider had simply measured the total radioactivity, using a scintillation counter, of a fraction that wasn't even pure cytosine—a fraction simply purified by extraction and centrifugal techniques.
Strange as it may sound, I don't think this transfer has even been proven. The more modern kinetic studies simply measure the sum of ·CH₃ transferred, assuming that they're going to carbon five as though a foregone conclusion. They never attempt to differentiate between 5-methylcytosine and N-methylcytosine, neither analytically or even conceptually.
In every single case, it appears, biochemists simply measure the total ¹⁴CH₃ transferred to DNA. There is no reason to assume, besides the enzyme's formal name, that these methyl groups are adding to cytosine's carbon number five. As if caught in a semantic web, the inappropriate name given to this enzyme forces them to make one big assumption: The enzyme called cytosine-5-methyltransferase selectively transfers methyl groups to cytosine's carbon five.
Rates of less than one methyl group transferred per second were measured.
This is actually slower that Na⁺/K⁺-ATPase, almost like it's not even trying.
"However, the distortion that occurs is as surprising as it is elegant: the m5C-MTases cleanly extend the target cytosine out of the helix and into the catalytic site, without seriously disturbing the rest of the DNA helix." —Kumar, Sanjay
By contrast, an enzyme known to methylate adenosine on the nitrogen is known operate over a million times faster (Reich, 1992).
Sneider, T. W., W. M. Teague, and L. M. Rogachevsky. "S-adenosylmethionine: DNA-cytosine 5-methyltransferase from a Novikoff rat hepatoma cell line." Nucleic acids research (1975)
Vilkaitis, Giedrius, et al. "The mechanism of dna cytosine-5 methylation kinetic and mutational dissection of hhai methyltransferase." Journal of Biological Chemistry (2001)
Bacolla, Albino, et al. "Recombinant human DNA (cytosine-5) methyltransferase II. Steady-state kinetics reveal allosteric activation by methylated DNA." Journal of Biological Chemistry (1999)
Gowher, Humaira, and Albert Jeltsch. "Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites." Journal of molecular biology 309.5 (2001): 1201-1208.
Reich, N. O., et al. "In vitro specificity of EcoRI DNA methyltransferase." Journal of Biological Chemistry (1992)
Kumar, Sanjay, et al. "The DNA (cytosine-5) methyltransferases." Nucleic acids research(1994)
XII. The CpG repeat
Carbon five-'methylated' cytosine (C) is found more often than next to a guanosine (G) on the DNA backbone—separated by a phosphate (p). So much so, in fact, that CpG islands are considered indicative of genes controlled by 'methylation,' and they are often taken as representative of DNA 'methylation' potential. This cannot be denied and needs to be stressed; this is of of prime importance.

The prevalence of CpG islands in DNA sequences should tell us whether or not said gene is controlled by methylation. These repeat is unidirectional; it's not synonymous with its reverse (CpG ≠ GpC). Randomly expected prevalence of such repeats is estimated at 4.41% of the genome, though you probably would have guessed 6.25% (¹⁄₄ × ¹⁄₄ = ¹⁄₁₆). [This is because cytosine and guanosine are found at a prevalence of ~21%, not 25%.] However, the actual prevalence of CpG sequences—in humans—is found to be ~1%.
Under the scheme that I am proposing, you would expect the genes that control aspartate–glutamate metabolism to be "CpG islands." Let us take a look at one common definition of the "CpG island."
The aforementioned island, the one for D-aspartate oxidase, certainly does fall under this definition. The first 1000·bp of this gene has a found/expected ratio of 29.4—the Australia of CpG islands (using 1% as the denominator). In contrast, the first 1,000·bp of the gene encoding fatty acid synthase has only five CpG repeats, giving it a CpG(f)/CpG(e) ratio of exactly one—the expected prevalence for CpG repeats.
Atkinson, Nigel. "Biology 327: Epigenetics." University of Texas—Austin
Anonymous Sequencer. "D-aspartate oxidase, exon 1-3." European Nucleotide Archive
Anonymous Sequencer. "Chicken fatty acid synthase gene." European Nucleotide Archive
XIII. Confirming the Hypothesis
The gene which encodes aspartate transaminase in Ralstonia solanacearum is listed as 1185·bp long. Of the first one thousand nucleotides, there are 135 CpG repeats. The gives this gene a found over expected ratio of 27. Since the gene which encodes D-aspartate oxidase is an established CpG island, this one must also be considered a CpG island.

The gene encoding γ-glutamyl transferase has a ratio of 29 for its first 1000·bp, higher than D-aspartate oxidase. This represents a 2900% enrichment over what you'd expect to find from a random sequence.
As the enzyme which creates β-methylaspartate, and thus 5-methylcytosine, you'd expect gluatamate mutase to enriched. This enzyme occupies a central hub in aspartate–glutamate metabolism. For this reason, I had decided to count the CpG repeats in the entire 4042·bp gene. I found 374 such repeats, for an absolute prevalence of 18.5%. The expected prevalence of 1% can even be considered generous in light of findings.
Anonymous Sequencer. "Ralstonia solanacearum Aspartate transaminase." European Nucleotide Archive
Anonymous Sequencer. "Streptomyces malaysiensis putative gamma-glutamyl transferase." European Nucleotide Archive
Anonymous Sequencer. "Citrobacter amalonaticus DNA for glutamate mutase." European Nucleotide Archive
Bird, Adrian P. "DNA methylation and the frequency of CpG in animal DNA." Nucleic acids research (1980)
XIV. The Methylaspartate Cycle
"The methylaspartate cycle branches off the tricarboxylic acid cycle on the level of 2-oxoglutarate. [...] It allows the separation of the flows of the methylaspartate and tricarboxylic acid cycles, thus preventing the competition between these two cycles for intermediates." ―Farshad Borjian
Holobacteria have much higher β-methylasparate than mammals. From this, you would expect it to have more 5-methylcytosine in its DNA.
This certainly appears to be the case. The nucleic cytosine of halobacteria is 100% 5-methylcytosine.
Borjian, Farshad, et al. "The methylaspartate cycle in haloarchaea and its possible role in carbon metabolism." The ISME journal (2016)
Vogelsang-Wenke, Heike. "Isolation of a halobacterial phage with a fully cytosine-methylated genome." Molecular and General Genetics (1988)
XV. Anatomy of a Unicorn
The enzyme usually assumed to create all the natural 5-methylcytosine on the planet has been purified, crystallized, and imaged. This was done despite it's extremely low catalytic rates, conversions which would likely approach that of S-adenosylmethionine and DNA—'enzyme' excluded. Nonetheless, this has been done. The structure of this enzyme has been characterized down to 2.8 angstroms, and even more bizarre reaction schemes had to be created to account for this increased resolution.
The magic base, usually just assumed to make an apparition at the right moment, was nowhere to be found.
So conspicuous was its absence that water was even hypothesized to assume this role—to actually be capable of extracting a hydrogen from an sp³-hybridized carbon. But thankfully, there was no evidence of this; so we don't have to think too hard about dissolving in rainstorms.
Glutamate needs to become glutamic acid for this scheme, which is difficult to imagine at bodily pH. This doesn't seem to matter however, since the interior of any enzyme can always be imagined as 'inaccessible.' But this contradicts their very scheme, since they need water to abstract the proton from carbon five.
This enzyme is presumed to work on double-stranded DNA by these authors, a fact which forces the reaction scheme from the impossible towards the absurd:
"M. Haelll provides an even more extreme example of DNA distortion." ―Karin M. Reinisch
The initial step, the incorporation of the target cytosine, is energetically impossible: A 'breathing' mechanism is actually proposed to account for this.
Reinisch, Karin M., et al. "The crystal structure of Haelll methyltransferase covalently complexed to DNA: An extrahelical cytosine and rearranged base pairing." Cell (1995)
XV. Lack of Evidence for Carbon–Carbon Methylation
There is a conspicuous lack of evidence that this enzyme can do as stated—that it can actually transfer a methyl group to the five carbon of cytosine.
XVI. Implications
The assumption that this 'enzyme' controls DNA methylation influences the conclusions made in hundreds of scientific articles. This assumption appears to have started in the years following its 'discovery,' and was highly influenced by Sneider having it named "DNA-cytosine 5-methyltransferase." He jumped the gun, made assumptions, and thought he'd found a way to explain 5-methylcytosine's existence in DNA. But any radioactive methyl groups were most likely simply transferred to the 4-nitrogen during those studies . . . and at snail-like pace.
There is no indication that the nuclear 'enzyme' called cytosine 5-methyltransferase is responsible for 5-methylcytosine's existence in DNA, and every indication that β-methylaspartate is. Consuming more glutamate than asparate would be expected to increase increase β-methylaspartate levels, 5-methylcytosine levels, and total DNA methylation—which is especially concentrated on the DNA regulating glutamate and aspartate metabolism, as an evolutionary control mechanism. The only strong correlation that will ever be found examining DNA methylation will probably be dietary glutamate/aspartate ratios and vitamin B₁₂—necessary for transforming glutamate into β-methylaspartate: the precursor for 5-methylcytosine, the presence of which determines DNA methylation as it's most often defined.
This post shows why vitamin B₁₂ and glutamate are the only things capable of significantly changing DNA methylation levels, why methionine and folate are ineffective, and points-out the absurdity of the modern paradigm.
The methylation of DNA is an undeniable phenomenon, and found in most living organisms. The enzymes responsible for this have been extracted, purified, assayed—with their kinetic rates determined, giving indisputable proof that DNA methylation can be an enzymatic event. The DNA encoding for such enzymes have been genetically sequenced, synthesized, and recombined to form fully-functional constructs . . . and the X-structures have been elucidated.
But there exists three forms of methylated DNA: oxygen-linked, as in the case of 6-O-methylguanidine; nitrogen-linked as in the case of N-6-methyladenosine, N-3-methylthymine, and N-4-methylcytosine; and surprisingly, carbon-linked as in the single case of C-5-methylcytosine. All of these modified nucleotides exist and can be found in DNA, and one of them has dubious origins—a biolochemical unicorn.
While this may seem trivial, it is not; as you will see. Misunderstandings surrounding the origin of 5-methylcytosine obfuscates epidemiological correlations, hides important cellular control mechanisms, and can perhaps can even lead to further misunderstandings if compounded—snowballed around the original misconception.
I. The Modern Paradigm
"Thereby, methylation adds extra information to the DNA that is not encoded in the sequence, and the methylated bases can be considered the 5th, 6th, and 7th letters of the genetic alphabet." —Albert Jeltsch
Most people have heard of DNA methylation, and had probably heard some speculations towards its purpose. There exists hundreds, if not thousands, of studies attempting to correlate DNA methylation with such things as: longevity, cancer, and diabetes—all with basically null results. Much paper has been wasted in this manner, with a small forest killed by attempting to explain a few classic rat–methionine studies in this way.
But a clear trend has emerged: Methylated DNA strands appear to be more stable; they are slighly-downregulated genes. The methylation of DNA is the protoypical epigenetic mechanism, and the only one acknowledged by many biologists.
"In higher eukaryotes DNA methylation is the only known covalent modification of the DNA." —Albert Jeltsch
These methylations are also thought to protect cellular DNA from its own enzymes—enzymes which could exist to destroy foreign DNA, or as simply a recycling measure. There are many other speculations on the function and purpose of DNA methylation, but a few things should perhaps be considered before entertaining some of the more abstract and tenable hypotheses out there.
Jeltsch, Albert. "Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases." Chembiochem (2002)
II. Why Carbon:Carbon Bonds are Different
The formation of the carbon:carbon bond is a relatively rare event in biochemistry—almost unheard-of without a carbonyl or phosphate involved. The electronegative oxygen pulls electrons from the carbonyl carbon, allowing backside attack from electron-rich nucleophiles such as a thiols and amides. The condensation reactions in steroid synthesis make use of phosphate groups on the terminal carbons, 'activating' them with electronegativity.
The DNA methyltransferase reaction—which catalyzes the carbon:carbon bond formation of 5-methylcytosine—involves adding a methyl group to a resonant ring. You'd be hard-pressed to find another instance of such, or even another biochemical carbon:carbon bond formation besides maybe the rare Diels–Alder.
The #5 bond of methylcytosine is the only variable carbon:carbon bond in DNA. All other methylations are to either to nitrogen or oxygen, additions which are in no way unusual: Such examples can be found in familiar molecules such as melatonin, methoxytyramine, and epinephrine. So when talking about 'DNA methylation' it becomes necessary to make the distinction between what atoms have been methylated, or assumed to have been.. .
"So far, in metazoa only cytosine-C5 methylation has been found in DNA; this methylation mainly occurs at CG sequences, about 60 ± 90% of which are modified in mammals (corresponding to 3 ± 8% of all cytosine residues)." —Albert Jeltsch
It was also the first one discovered.
Jeltsch, Albert. "Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases." Chembiochem (2002).
III. Help from DNA Methyltransferase
"Although AdoMet is a very effective donor for methyl groups, methylation of cytosine residues at position 5 is not a trivial reaction." —Albert Jeltsch
The enzyme DNA methyltransferase is thought to do what no other can. It breaks the resonance of the pyrimidine ring with a protonated glutamyl, while forming a thioether with carbon four of cytosine. You might be tempted to wonder why this is so, but you wouldn't want to since it doesn't happen.
"DNA (cytosine-5) methyltransferases introduce a methyl group onto carbon 5 of the target cytosine through a covalent intermediate between the protein and the target cytosine. During this process, the cytosine is flipped 180° out of the DNA backbone into an active site pocket of the enzyme." ―Sriharsa Pradham
Carbon five—the site of the imaginary reaction—actually faces the DNA backbone. The enzyme is imagined to work on single-stranded DNA: It is also thought to perform miracles.
"Therefore, the catalytic mechanism of C-MTases can explain why DNA MTases developed a base-flipping mechanism." ―Jeltsch
"The nature of the proton abstracting base is not known with certainty," —Albert Jeltsch
This is a base later found to not exist at 2.8 Å resolution.(Reinisch, 1995).
Of course all of this is too small to see, so they can just sprinkle fairy dust to obscure any irregularities. Step four is technically the molecular equivalent of Bruce Lee in "The Way of the Dragon."
"Although these studies were carried out with different enzymes and none of them finally proves that a base rotating mechanism is operative." —Albert Jeltsch
Jeltsch, Albert. "Beyond Watson and Crick: DNA methylation and molecular enzymology of DNA methyltransferases." Chembiochem (2002)
IV. Failure of S-Adenosylmethionine to Increase Methylation"Although MET supplementation significantly decreased the [SAM]:[SAH] ratio in liver and brain, no significant dietary effects on genome-wide DNA methylation were found." —Robert A. Waterland
All so-called DNA methyltransferase enzymes use S-adenosylmethionine as cofactor. One would then expect this cofactor to increase total DNA methylation by ingestion or by injection. It does, after all, the source of the methyl groups in question.
"Methionine supplementation increased the renal concentration of SAH without changing the SAM/SAH ratio. This unchanged profile was also observed for DNA methylation at the promoter region of the p53 gene." —Cátia Lira DoAmaral
The failure of studies to demonstrate changes in 'DNA methylation'—as defined as 5-methylcytosine—by methionine supplementation shouldn't go unnoticed. It's unfortunate that many researchers fail to differentiate between the possible and the impossible methylation events in titles and abstracts, as this only adds confusion to an already confused paradigm.
Waterland, Robert A. "Assessing the effects of high methionine intake on DNA methylation." The Journal of nutrition (2006)
Do Amaral, Cátia Lira. "The effects of dietary supplementation of methionine on genomic stability and p53 gene promoter methylation in rats." Mutation Research/Genetic Toxicology and Environmental Mutagenesis (2011)
V. Failure of Folate to do Likewise
"...however, there was not a consistent association between global DNA methylation and folate status across studies." —Krista S. Crider
Although not considered a cofactor for the DNA methyltransferase reactions, folate is dragged into this imply because it regenerates S-adenosylmethionine. But if S-adenosylmethionine itself cannot even influence DNA methylation, how can you folate capable of this?
When measuring the effects of folate on hundreds of people, the results were negligible—basically null.
"Women with higher (vs. lower) RBC folate had higher mean DNA methylation (5.12 vs. 4.99%) in the pre-fortification period, but lower (4.95 vs. 5.16%) DNA methylation in the post-fortification period." —Sajin Bae
This was defined with respect to 5-methylcytosine, or course. Increases in nitrogen-methylated DNA is certainly possible from supplemental methionine, but N-methylation is rarely studied.
"Global DNA methylation was assessed by liquid chromatography-tandem mass spectrometry and expressed as a percentage of total cytosine." —Sajin Bae
Should anyone think that a .03% change is something to consider, take a look at what vitamin B₁₂ can do. It can cause great changes in spite of the fact that it's not a cofactor for DNA methyltransferse.
Crider, Krista S. "Folate and DNA methylation: a review of molecular mechanisms and the evidence for folate’s role." Advances in Nutrition: An International Review Journal (2012)
Bae, Sajin, et al. "Impact of folic acid fortification on global DNA methylation and one-carbon biomarkers in the Women's Health Initiative Observational Study cohort." Epigenetics (2014)
VI. Vitamin B₁₂ Increases Methylation—and Nothing Else
"After 10 wk, the colonic DNA of the deficient rats displayed a 35% decrease in genomic methylation..." —Sang-Woon Choi
The only thing shown to reliably change DNA methylation has been vitamin B₁₂. Nobody seems to mention this fact, a fact which conflicts with the commonly held notion that the S-adenosylmethionine—and anything that re-methylates it—will do this. Folate and choline could perhaps be imagined as almost forcing methyl groups onto DNA by positive pressure in the methyl pool: two hydraulic analogies with no place in the genesis of 5-methylcytosine—although perhaps appropriate when talking about catecholamines and homocysteine.
"Identification of cytosine and 5-methylcytosine was obtained by MS analysis of chromatographic peaks." —Sang-Woon Choi
By unambiguous methods: Vitamin B₁₂ had been shown to increase cytosine methylation; something which cannot be achieved reliably by either folic acid or S-adenosylmethionine—the definitive methyl donor, making this especially enigmatic since S-adenosylmethionine is the very cofactor for the enzyme thought responsible.
Choi, Sang-Woon, et al. "Vitamin B-12 deficiency induces anomalies of base substitution and methylation in the DNA of rat colonic epithelium." The Journal of nutrition (2004)
VII. How 5-Methylcytosine is Actually Formed
Regular cytosine is formed from the amino acid aspartate. This is a straghtforward process, and had been known since the '50s.
A small metabolite called carbamoyl phosphate is attacked by the amide (shown below as an amine but the hydrogen is removed by the enzyme) of aspartate, held in position and acted-upon by the enzyme carbamoyl transferase. This is not a carbon:carbon addition, and neither is the final condensation stage. Such interactions are known to occur all the time.
But when β-methylaspartate is acted upon by these enzymes, you would expect the 5-methylcytosine precursor to be formed: 5-methyldihydroororate, or 5-methyldihydroororic acid (depending on pH.)
Which would then go on to become 5-methylcytidine, and then incorporated into DNA. The substrate β-methylaspartate is no stranger to biology: This molecule is formed through the vitamin B₁₂-catalyzed enzyme glutamate mutase—named so because it actually isomerizes glutamate into β-methylaspartate, similar to B₁₂'s ability to isomerize methylmalonic acid.
 Vitamin B₁₂ is only thing capable of isomerizing glutamate into β-methylaspartate.
Vitamin B₁₂ is only thing capable of isomerizing glutamate into β-methylaspartate.Now this is the formation of a carbon:carbon bond, to be sure, and could appear no less difficult than the one in question: the methlyation of cytosine on single-stranded DNA. But keep-in-mind that vitamin B₁₂, or cobalamin, is a giant heme-like ring structure with a cobalt atom in the centre. This gives it high electric potential, something that the DNA methyltransferase or S-adenosylmethionine simply doesn't have.
When the glutamate/aspartate ratio is high, you might expect more glutamate to be be converted to β-methylaspartate through the action of glutamate mutase. This is the precursor the 5-methylcytosine, and has nothing do with methionine.
Yates, Richard A. "Pyrimidine biosynthesis in Escherichia coli." Journal of Biological Chemistry (1956)
VIII. Confirmation from Neurotransmitter Studies
"Our data suggest that DNA methylation status controls transcription-dependent regulation of glutamatergic synaptic homeostasis." —Jarrod P. Meadows
You might also expect this to happen anywhere . . . even in the brain, the place most suitable for such an investigation. The brain has a highly-variable glutamate/aspartate ratio, and thus is a prime candidate for such analysis. These two amino acids are considered to be neurotransmitters involved in learning, motivation, and memory.
"For this reason, many years ago, Francis Crick proposed that a self-perpetuating biochemical autoconversion of methylated DNA might serve as a memory mechanism at the molecular level." —Jarrod P. Meadows
Following in the footsteps of Francis Crick, Jarrod Meadows had decided to investigate the effect of glutamate on DNA methylation in the brain. He used tetrodotoxin, as this is the standard molecule used to inhibit glutamate release in the brain. The low brain glutamate was confirmed, and he had found that this had resulted in DNA methylation changes greater than those resulting from inhibiting DNA methyltransferase. Tetrodotoxin, ostensibly through glutamate, did this in spite of contributing no methyl groups whatsoever.
"Inhibiting neuronal activity with tetrodotoxin decreased the cytosine methylation of and increased the expression of genes encoding glutamate receptors and trafficking proteins," —Jarrod P. Meadows
Decreasing glutamate would inhibit the formation of β-methylaspartate, the precursor for 5-methylcytosine, through glutamate mutase. This would lead to lower DNA methylation, exactly as was found.
Other studies have confirmed such a relationship, between asparate levels and DNA methylation (Punzo, 2016).
In the Meadow's study: The genes hypomethylated from low glutamate were, and perhaps not surprisingly, involved in aspartate signalling:
"An additional series of studies found that the Bdnf gene locus is also subject to memory-associated changes in DNA methylation and that this effect is regulated by the N-methyl-D-aspartate (NMDA) receptor," —Jarrod P. Meadows
And also in glutamate signalling:
"One particularly interesting possibility is that regulation of methylation of the genes for AMPA-subtype glutamate receptors might be involved, as well as methylation of those genes regulating their trafficking. For example, Jayanthi et al. have observed activity-induced alterations in AMPA receptor gene methylation that correlate with glutamate receptor expression in vivo. Previous studies as well as our own data have demonstrated that TTX-induced synaptic scaling is associated with altered glutamate receptor gene transcription as well as that of arc, an AMPA receptor trafficking regulator." —Jarrod P. Meadows
This strengthens the idea that the glutamate/aspartate ratio is controlling DNA 'methylation' on the very genes responsible for its regulation—having nothing to do with methionine. This might seem paradoxical, if not for the facts outlined above. With these in mind, this all makes perfect sense.
Meadows, Jarrod P., et al. "DNA methylation regulates neuronal glutamatergic synaptic scaling." Science signaling (2015)
Punzo, Daniela, et al. "Age-related changes in D-aspartate oxidase promoter methylation control..." Journal of Neuroscience (2016)
IX. Hints from Genetics
The methylation of DNA—as defined by 5-methylcytosine—is routinely measured on select genes. This is because: Some regions are particularly-rich in cytosine, leading to more accurate measurements in these places. One of the genes commonly tested for methylation happens to be the one encoding for D-aspartate oxidase:
"The choice of Ddo as a model gene was mainly based on the facts that this gene is developmentally regulated in brain by DNA methylation changes. Age-Related changes in D-aspartate oxidase promoter methylation control extracellular D-aspartate levels and prevent precocious cell death during brain aging. The average methylation levels range between 60 and 30% in different stages." —Ermanno Florio
You would then think that highly-'methylated' areas would be found in the genes responsible for controlling glutamate:aspartate metabolism, including—but not limited to—aspartate transaminase, γ-glutamyl transferase, and glutamyl mutase.
Florio, Ermanno, et al. "Tracking the evolution of epialleles during neural differentiation and brain development: D-Aspartate oxidase as a model gene." Epigenetics (2017)
X. Five-Methycytosine
"In Fig. 2 a minor constituent designated “epicytosine” is indicated, having a migration rate somewhat greater than that of cytosine." ―Rollin D Hotchkiss
Was the sixth nucleotide discovered, and the first assumed to be a post-translational modification. Originally discovererd in 1925, it's existence hadn't been confirmed until 1948. Two years later its existence was headlined in the high-visibility journal Nature.
"...nevertheless, the fraction is distinct from cytosine and is clearly not uracil." ―Rollin D Hotchkiss
Five-methylcytosine was on the minds of researchers then, and the nitrogen-methylated nucleotides weren't known to exist until later.
Johnson & Coghill. "Researches on pyrimidines. The discovery of 5-methyl-cytosine in tuberculinic acid..." Journal of the American Chemical Society (1925)
Hotchkiss, Rollin D. "The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography." Journal of Biological Chemistry (1948)
Wyatt, G. R. "Occurrence of 5-methyl-cytosine in nucleic acids." Nature (1950)
XI. DNA Methyltransferase
"The rate of methylation observed in this study (31 pmoles/mg DNA/hour), although 85-fold higher than the rate using DNA from mid-log phase cells, is low compared to the calculated pmoles of methyl group needed for saturation of DNA completely devoid of methyl groups which is calculated to be on the order of 15,000 pmoles/ mg DNA." —T.W. Sneider
With 5-methylcytosine the only known 'DNA methylation' product, any transfer of methyl groups measured from S-adenosylmethionine to DNA would simply been assumed to be those forming 5-methylcytosine. The variable nature of 5-methylcytosine content found in DNA further would further confound such measurements, turning the negligible enzymatic rates into something even less.
Moreover, Sneider was likely measuring the sum of all of the DNA methytransferase rates combined, even the real ones. This is a possibility that he'd even mentioned himself, a likelihood which would further marginalize the already snail-like catalytic rates. He also didn't prove that the methyl groups were actually transferred to cytosine at carbon five; Sneider had simply measured the total radioactivity, using a scintillation counter, of a fraction that wasn't even pure cytosine—a fraction simply purified by extraction and centrifugal techniques.
Strange as it may sound, I don't think this transfer has even been proven. The more modern kinetic studies simply measure the sum of ·CH₃ transferred, assuming that they're going to carbon five as though a foregone conclusion. They never attempt to differentiate between 5-methylcytosine and N-methylcytosine, neither analytically or even conceptually.
"The measured methylation rate constant kchem = 0.26 s⁻¹ for WT M.HhaI agrees well with that recently reported." —Giedrius Vilkaitis
In every single case, it appears, biochemists simply measure the total ¹⁴CH₃ transferred to DNA. There is no reason to assume, besides the enzyme's formal name, that these methyl groups are adding to cytosine's carbon number five. As if caught in a semantic web, the inappropriate name given to this enzyme forces them to make one big assumption: The enzyme called cytosine-5-methyltransferase selectively transfers methyl groups to cytosine's carbon five.
"The turnover number for the enzyme varied considerably among the DNA templates, from ∼1 to 50 h⁻¹..." —Albino Bacolla
Rates of less than one methyl group transferred per second were measured.
"With oligonucleotide substrates, the catalytic activity of Dnmt3a is similar to that of Dnmt1: the Km values for the unmethylated and hemimethylated oligonucleotide substrates are 2.5 μM, and the kcat values are 0.05 h⁻¹ and 0.07 h⁻¹, respectively." —Albert Jeltsch
This is actually slower that Na⁺/K⁺-ATPase, almost like it's not even trying.
"However, the distortion that occurs is as surprising as it is elegant: the m5C-MTases cleanly extend the target cytosine out of the helix and into the catalytic site, without seriously disturbing the rest of the DNA helix." —Kumar, Sanjay
By contrast, an enzyme known to methylate adenosine on the nitrogen is known operate over a million times faster (Reich, 1992).
Sneider, T. W., W. M. Teague, and L. M. Rogachevsky. "S-adenosylmethionine: DNA-cytosine 5-methyltransferase from a Novikoff rat hepatoma cell line." Nucleic acids research (1975)
Vilkaitis, Giedrius, et al. "The mechanism of dna cytosine-5 methylation kinetic and mutational dissection of hhai methyltransferase." Journal of Biological Chemistry (2001)
Bacolla, Albino, et al. "Recombinant human DNA (cytosine-5) methyltransferase II. Steady-state kinetics reveal allosteric activation by methylated DNA." Journal of Biological Chemistry (1999)
Gowher, Humaira, and Albert Jeltsch. "Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpA sites." Journal of molecular biology 309.5 (2001): 1201-1208.
Reich, N. O., et al. "In vitro specificity of EcoRI DNA methyltransferase." Journal of Biological Chemistry (1992)
Kumar, Sanjay, et al. "The DNA (cytosine-5) methyltransferases." Nucleic acids research(1994)
XII. The CpG repeat
Carbon five-'methylated' cytosine (C) is found more often than next to a guanosine (G) on the DNA backbone—separated by a phosphate (p). So much so, in fact, that CpG islands are considered indicative of genes controlled by 'methylation,' and they are often taken as representative of DNA 'methylation' potential. This cannot be denied and needs to be stressed; this is of of prime importance.
The prevalence of CpG islands in DNA sequences should tell us whether or not said gene is controlled by methylation. These repeat is unidirectional; it's not synonymous with its reverse (CpG ≠ GpC). Randomly expected prevalence of such repeats is estimated at 4.41% of the genome, though you probably would have guessed 6.25% (¹⁄₄ × ¹⁄₄ = ¹⁄₁₆). [This is because cytosine and guanosine are found at a prevalence of ~21%, not 25%.] However, the actual prevalence of CpG sequences—in humans—is found to be ~1%.
"...the frequency of CpG is only 20% of the predicted value." ―Nigel Atkinson
Under the scheme that I am proposing, you would expect the genes that control aspartate–glutamate metabolism to be "CpG islands." Let us take a look at one common definition of the "CpG island."
"Q: How is it defined? A: 200-3000 bp in length, and greater than 60% CpG [relative to expected value]." ―Nigel Atkinson
The aforementioned island, the one for D-aspartate oxidase, certainly does fall under this definition. The first 1000·bp of this gene has a found/expected ratio of 29.4—the Australia of CpG islands (using 1% as the denominator). In contrast, the first 1,000·bp of the gene encoding fatty acid synthase has only five CpG repeats, giving it a CpG(f)/CpG(e) ratio of exactly one—the expected prevalence for CpG repeats.
Atkinson, Nigel. "Biology 327: Epigenetics." University of Texas—Austin
Anonymous Sequencer. "D-aspartate oxidase, exon 1-3." European Nucleotide Archive
Anonymous Sequencer. "Chicken fatty acid synthase gene." European Nucleotide Archive
XIII. Confirming the Hypothesis
The gene which encodes aspartate transaminase in Ralstonia solanacearum is listed as 1185·bp long. Of the first one thousand nucleotides, there are 135 CpG repeats. The gives this gene a found over expected ratio of 27. Since the gene which encodes D-aspartate oxidase is an established CpG island, this one must also be considered a CpG island.
The gene encoding γ-glutamyl transferase has a ratio of 29 for its first 1000·bp, higher than D-aspartate oxidase. This represents a 2900% enrichment over what you'd expect to find from a random sequence.
As the enzyme which creates β-methylaspartate, and thus 5-methylcytosine, you'd expect gluatamate mutase to enriched. This enzyme occupies a central hub in aspartate–glutamate metabolism. For this reason, I had decided to count the CpG repeats in the entire 4042·bp gene. I found 374 such repeats, for an absolute prevalence of 18.5%. The expected prevalence of 1% can even be considered generous in light of findings.
"Thus in human DNA, where the fraction of (G+C) is 0.4, we would expect CpG to occur with a frequency of 0.2 x 0.2 = 0.04, whereas the observed frequency is about 0.008. [.8%]" ―Adrian P.Bird
Anonymous Sequencer. "Ralstonia solanacearum Aspartate transaminase." European Nucleotide Archive
Anonymous Sequencer. "Streptomyces malaysiensis putative gamma-glutamyl transferase." European Nucleotide Archive
Anonymous Sequencer. "Citrobacter amalonaticus DNA for glutamate mutase." European Nucleotide Archive
Bird, Adrian P. "DNA methylation and the frequency of CpG in animal DNA." Nucleic acids research (1980)
XIV. The Methylaspartate Cycle
"Haloarchaea (class Halobacteria) live in extremely halophilic conditions and evolved many unique metabolic features, which help them to adapt to their environment. [...] Aerobic haloarchaea gained two anaplerotic acetate assimilation pathways, the glyoxylate cycle and the methylaspartate cycle." ―Farshad Borjian
Halobacteria have a methylasparate cycle. This is a metabolic cycle; a branch-off of the Citric Acid Cycle where glutamate is isomerized by glutamate mutase, deaminated, and used for energy.
"The methylaspartate cycle branches off the tricarboxylic acid cycle on the level of 2-oxoglutarate. [...] It allows the separation of the flows of the methylaspartate and tricarboxylic acid cycles, thus preventing the competition between these two cycles for intermediates." ―Farshad Borjian
This certainly appears to be the case. The nucleic cytosine of halobacteria is 100% 5-methylcytosine.
"The genome of ΦN consists of linear double-stranded DNA, 56 kb in size, whose dCMP is totally replaced by 5-methyl-dCMP. This is the second case of a fully cytosine-methylated genome..." ―Heike Vogelsang-Wenke
More indication that DNA 'methylation' is simply a function of β-methylasparate levels (C-methylation).
"Therefore, the methylaspartate cycle appears to be a glutamate overflow mechanism: it functions only at glutamate concentrations, which exceed a certain threshold signalizing the availability of nitrogen and energy to perform anabolic reactions." ―Farshad Borjian
Borjian, Farshad, et al. "The methylaspartate cycle in haloarchaea and its possible role in carbon metabolism." The ISME journal (2016)
Vogelsang-Wenke, Heike. "Isolation of a halobacterial phage with a fully cytosine-methylated genome." Molecular and General Genetics (1988)
XV. Anatomy of a Unicorn
The enzyme usually assumed to create all the natural 5-methylcytosine on the planet has been purified, crystallized, and imaged. This was done despite it's extremely low catalytic rates, conversions which would likely approach that of S-adenosylmethionine and DNA—'enzyme' excluded. Nonetheless, this has been done. The structure of this enzyme has been characterized down to 2.8 angstroms, and even more bizarre reaction schemes had to be created to account for this increased resolution.
"The basic moiety that abstracts the C5-H proton is not obvious from either complex." ―Karin M. Reinisch
The magic base, usually just assumed to make an apparition at the right moment, was nowhere to be found.
"Alternatively, a water molecule could act as the catalytic base. If so, there is no evidence for it in the M. Haelll electron density maps, but this may simply be a consequence of limited resolution (2.6 Å)." ―Karin M. Reinisch
So conspicuous was its absence that water was even hypothesized to assume this role—to actually be capable of extracting a hydrogen from an sp³-hybridized carbon. But thankfully, there was no evidence of this; so we don't have to think too hard about dissolving in rainstorms.
"a carboxyl oxygen of Glu-109 is hydrogen-bonded to the cytosine N3 at near neutral pH, the carboxyl pKa must be shlfted relative to the solution value of approximately 4.5. This shift, assumed in the proposed catalytic mechanism, is plausible, because Glu-109 is relatively solvent inaccessible." ―Karin M. Reinisch
Glutamate needs to become glutamic acid for this scheme, which is difficult to imagine at bodily pH. This doesn't seem to matter however, since the interior of any enzyme can always be imagined as 'inaccessible.' But this contradicts their very scheme, since they need water to abstract the proton from carbon five.
"the base pairing rearrangement required by M. Haelll is not energetically feasible." ―Karin M. Reinisch
This enzyme is presumed to work on double-stranded DNA by these authors, a fact which forces the reaction scheme from the impossible towards the absurd:
"M. Haelll provides an even more extreme example of DNA distortion." ―Karin M. Reinisch
"The lifetime of a GC pair in B-DNA is consistent with the relatively slow turnover rate of methyltransferases (e.g., 0.02/s for M. Hhal). Thus, the enzyme could capture the DNA as it breathes. However, the additional base rearrangements in M. Haelll are almost certain to be energetically costly, since the change in base pairing requires unstacking the DNA both opposite Gl 1’ and between pairs 9-9’ and 1 l-10’." ―Karin M. Reinisch
Is there anything this enzyme can't do?
"Second, it is more plausible that the substrate cytosine is flipped out as the DNA breathes." ―Karin M. Reinisch
A more plausible mechanism for the creation of 5-methylcytosine is through β-methylaspartate, before it's incorporated into DNA.
Reinisch, Karin M., et al. "The crystal structure of Haelll methyltransferase covalently complexed to DNA: An extrahelical cytosine and rearranged base pairing." Cell (1995)
XV. Lack of Evidence for Carbon–Carbon Methylation
There is a conspicuous lack of evidence that this enzyme can do as stated—that it can actually transfer a methyl group to the five carbon of cytosine.
XVI. Implications
The assumption that this 'enzyme' controls DNA methylation influences the conclusions made in hundreds of scientific articles. This assumption appears to have started in the years following its 'discovery,' and was highly influenced by Sneider having it named "DNA-cytosine 5-methyltransferase." He jumped the gun, made assumptions, and thought he'd found a way to explain 5-methylcytosine's existence in DNA. But any radioactive methyl groups were most likely simply transferred to the 4-nitrogen during those studies . . . and at snail-like pace.
There is no indication that the nuclear 'enzyme' called cytosine 5-methyltransferase is responsible for 5-methylcytosine's existence in DNA, and every indication that β-methylaspartate is. Consuming more glutamate than asparate would be expected to increase increase β-methylaspartate levels, 5-methylcytosine levels, and total DNA methylation—which is especially concentrated on the DNA regulating glutamate and aspartate metabolism, as an evolutionary control mechanism. The only strong correlation that will ever be found examining DNA methylation will probably be dietary glutamate/aspartate ratios and vitamin B₁₂—necessary for transforming glutamate into β-methylaspartate: the precursor for 5-methylcytosine, the presence of which determines DNA methylation as it's most often defined.
Last edited: